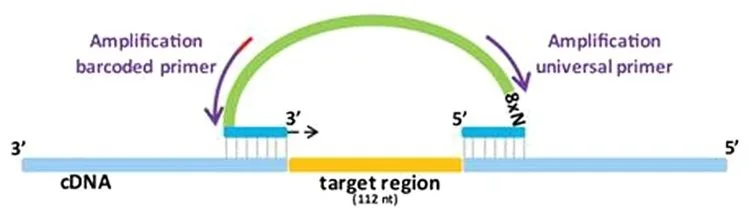
smMIP design
At TMP we are excited to bring the world of the microbiome to you. Using our smMIP technology platform, we can provide a targeted way to measure the microbiome in your sample, with a world of potential from research to health to agri- and food. Any previously characterized micro-organism can be detected by our targeted sequencing technology, without needing complicated metagenome sequencing. Simply pick what you wish to measure beforehand! Our pre-made panels can be used to measure several interesting pathways without any required customization.
Using our technology platform, TMP is able to design single-molecule molecular inversion probes (smMIPs) which target the gene- or genomic sequence that you wish to measure. smMIPs can be designed against almost any location in a microbial genome, allowing us to measure the regions and organisms you are interested in, leaving uninteresting data out. In combination with our high-throughput library prep methods, we can provide you the detail of metagenome sequencing for a 16S sequencing price.
smMIP design
smMIPs are created from a known sequence in a similar way to a PCR primer. A smMIP will have the ability to bind the desired target using its 2 hybridisation arms called the extension (ext) and ligation (lig) arm, which are connected by a structural backbone.
To find the best smMIPs for the region / gene, all possible smMIPs are created and a automated review process filters for the best predicted smMIPs. This is based on an in silico PCR analysis which evaluates where a smMIP might bind. The system looks at several smMIP characteristics to score them, including:
Hybridisation efficiency of extension and ligation arms
Species & Genus specificity
The number of genes / genomes targeted
The number of different genes / genomes targeted
The length of the target sequences
This leads to the selection of smMIPs which perform best across all categories.
High throughput, highly multiplexed sequencing
Once smMIPs are designed, we can apply them to capture the (c)DNA samples which you send us. The smMIPs hybridise to their target regions, after which a polymerase extends the extension arm, the smMIP is then ligated closed. The circular smMIPs are purified and converted to a library. Only captured sequences (the ones that we are interested in) end up in the library. This makes the signal to noise ratio very high as background (c)DNA is removed. Effectively, we enrich for the library reads we want, instead of trying to find them amongst reads we were never interested in. We also can achieve high coverage in a cost effective way. Many sample libraries created in this way are barcoded, pooled and massively parallel sequenced using Illumina sequencing technology. The result is then analysed and communicated back to you.